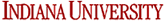Solvent suppression is needed when the solvent contains non-deuterated proton atoms, because the solvent signal could be over 1000 times stronger comparing to other signals of interest. It is a common technique for protein NMR since protein samples usually dissolve in 90%H2O/10%D2O solution instead of 100% D2O in order to investigate exchangble protons such as amide groups. In this case water suppression is absolutely necessary. On the other hand, the introduction of cold probes brings 3-4 times sensitivity gain over room temperature probes. Although it is great for some precious dilute samples, solvent suppression becomes more difficult with cold probes.
Below are some water suppression data collected on the standard ubiquitin sample using the 600 MHz equipped with a cold probe. The data were collected at 298 K.
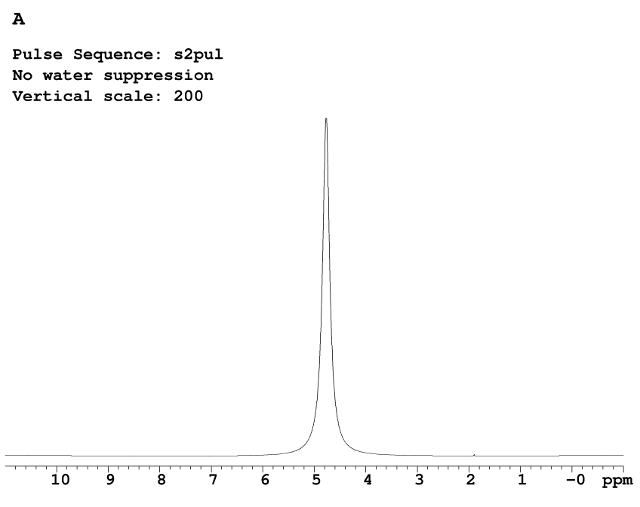
Figure A is the 1D 1H without any water suppression, The huge water signal at 4.76 ppm dominates the entire spectral region. Figures B, C, and D were collected with three different water suppression techniques.
Presatuation was used in Figure B. A low power was applied on water frequency during the relaxation delay period. The data after presat doesn’t look good. The water signal in this ubiquitin sample is very broad. The half-height linewidth is 95 Hz. The presat usually works great if the peak to be presaturated is sharp and narrow.
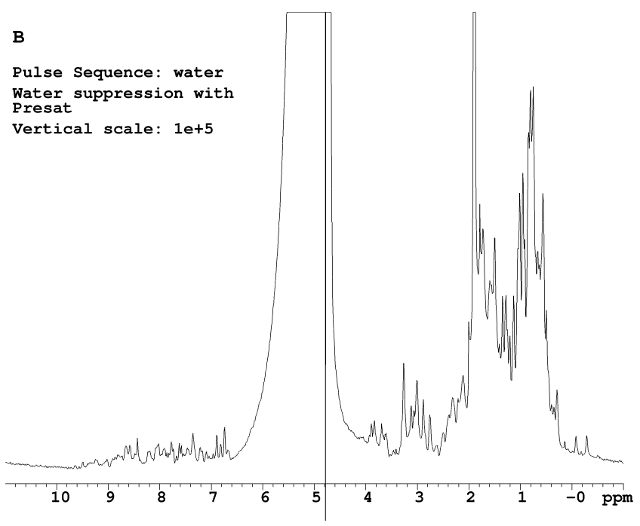
Watergate was used in Figure C. This technique usually works well on a room temperature probe. In this case, the water peak fluctuates abruptly probably due to radiation damping caused by the high sensitivity cold probe receiver coil.
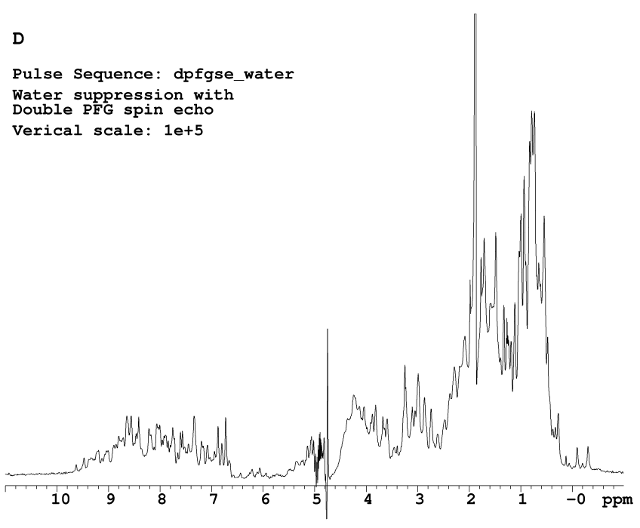
Figure D shows the best water suppression result using double PFG spin echo(dpfgse) sequence.
The pulse sequences used above are called water (for B and C), and dpfgse_water (for D) in VnmrJ software. Which one to choose really depends on the samples. Presat is good enough to suppress a sharp narrow signal, while dpfgse is more efficient for dilute macromolecular samples. Although the sequence is named ‘water’ or ‘dpfgse_water’, they can also be used for suppression of any other solvent signals. Water just happens to be the most common solvent for macromolecular NMR samples.
The default parameter values from VnmrJ template work pretty well, but It’s recommended users to spend a few minutes to optimize some of the most critical parameters. For example, in dpfgse_water sequence, you can ‘fine tune’ the solvent selective pulse power level ‘wrefpwrf‘. The default value given from the template is 2048. The easiest way is using array. Issuing the following on the command line,
array(‘wrefpwrf’,8,2000,50)
The above command would do a series of 1D using 8 wrefpwrf values starting with 2000, and increasing 50 each step. When it’s done, the following command wft dssh dssl would process and display the result side by side similar to the figure shown here. It is obvious that 2200 gives the best solvent suppression:
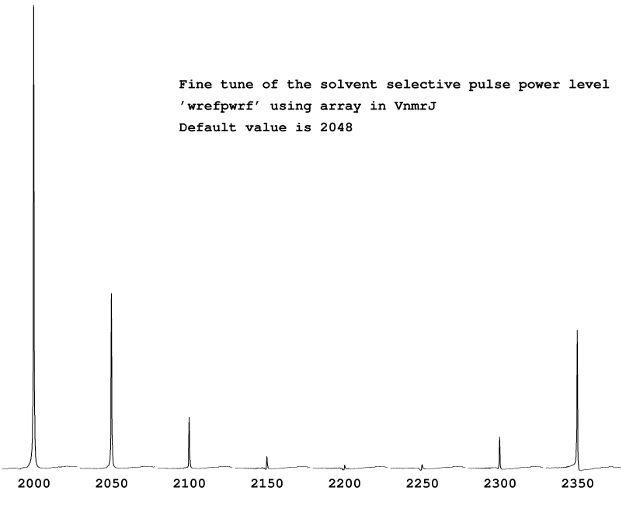
For the solvent suppression, one can array the saturation frequency, and power level to achieve the best result. Using array to check critical parameters is a good practice in order to get NMR data with the best resolution and sensitivity. This applies to not only 1D solvent suppression experiment, but any other NMR experiments as well.